HPC cluster
General information :
this equipment contributes to the rational design, supported by modeling, of radiopharmaceuticals labeled with innovative radionuclides.
The scientific software available on the cluster allows:
- to predict the structure and properties of molecules (bond strength, oxidation potential, etc.) by quantum chemistry calculations
- to model the ligand-biological receptor or protein-protein interactions with increasing degrees of sophistication (docking, molecular dynamics, QM/MM).
Principle of operation:

After establishing a secure connection to the service node, the user can run simulations in parallel mode (OpenMP or MPI) on up to 40 cores and a maximum elapsed time of 2000 hours. The simulations are scheduled by a management system, which is based on different submission queues. Execution starts according to the priorities of the selected queue and the previous use of cluster resources.
Specifications:
- 9 cluster nodes Dell R620 dual-core E5-2670 with 128 Go of RAM and 4 hard drives (RAID 0) at 15000 rpm
- 3 cluster nodes Dell R630 quad-core E5-2693 with 192 Go of RAM and 8 hard drives (RAID 0) at 15000 rpm
- Intel Infiniband switch
- molecular modeling softwares: Gaussian 16, TURBOMOLE 7.3, Molpro 2015.1, Schrödinger suite 2018, NWChem 6.6
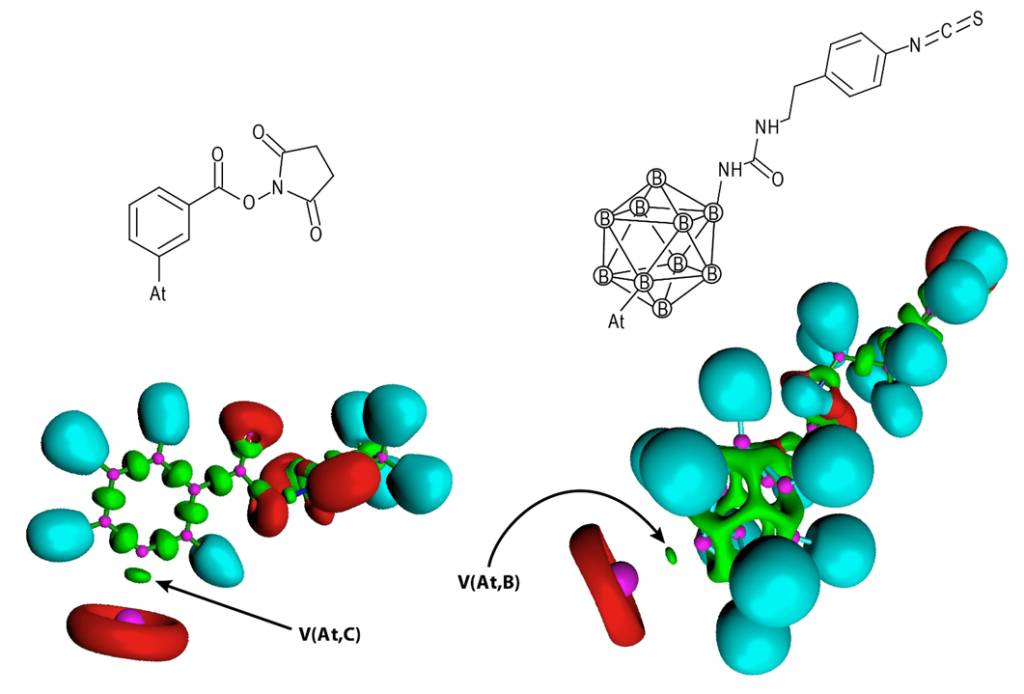 Figure 1 : 2D structures and representation of electron localization function (ELF) for two 211At-labeled compounds that have been evaluated in vivo.
Figure 1 : 2D structures and representation of electron localization function (ELF) for two 211At-labeled compounds that have been evaluated in vivo.
| Identity card | ||
| Name | PEKKA cluster | |
| Manufacturer | DELL | |
| ID number (internal ref.) | ||
| Place | CEISAM | |
| Owner | CEISAM | |
| Type og equipment | High performance computing cluster | |
| Information | ||
| Availability | Available | |
| Controlled zone | Delocalized access | |
| Restrictions on use | Training is needed | |
| Technical correspondent | Contact us | |
Additional information:
This equipment is only accessible:
- to molecular modeling specialists,
- after having completed training in the HPC cluster operating system.
Potential users are staff either from partners or from collaborative external academic teams.

